-
Notifications
You must be signed in to change notification settings - Fork 542
Optim-wip: Add main Activation Atlas tutorial & functions #782
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Changes from all commits
b7333ec
dae9576
f35b844
340d3b8
86eb581
f52ae2e
d51ef0d
6ac980c
18b017f
0233680
f8aa611
5b52643
e3c3457
dcf3b99
2e4f4b0
d1e22ae
271d845
ad03e3b
9b87f23
4a16d1a
29d0283
File filter
Filter by extension
Conversations
Jump to
Diff view
Diff view
There are no files selected for viewing
| Original file line number | Diff line number | Diff line change |
|---|---|---|
| @@ -1,7 +1,7 @@ | ||
| import functools | ||
| import operator | ||
| from abc import ABC, abstractmethod, abstractproperty | ||
| from typing import Any, Callable, Optional, Tuple, Union | ||
| from typing import Any, Callable, List, Optional, Tuple, Union | ||
|
|
||
| import torch | ||
| import torch.nn as nn | ||
|
|
@@ -27,7 +27,7 @@ def __init__(self) -> None: | |
| super(Loss, self).__init__() | ||
|
|
||
| @abstractproperty | ||
| def target(self) -> nn.Module: | ||
| def target(self) -> Union[nn.Module, List[nn.Module]]: | ||
| pass | ||
|
|
||
| @abstractmethod | ||
|
|
@@ -140,7 +140,9 @@ def loss_fn(module: ModuleOutputMapping) -> torch.Tensor: | |
|
|
||
| class BaseLoss(Loss): | ||
| def __init__( | ||
| self, target: nn.Module = [], batch_index: Optional[int] = None | ||
| self, | ||
| target: Union[nn.Module, List[nn.Module]] = [], | ||
| batch_index: Optional[int] = None, | ||
| ) -> None: | ||
| super(BaseLoss, self).__init__() | ||
| self._target = target | ||
|
|
@@ -150,7 +152,7 @@ def __init__( | |
| self._batch_index = (batch_index, batch_index + 1) | ||
|
|
||
| @property | ||
| def target(self) -> nn.Module: | ||
| def target(self) -> Union[nn.Module, List[nn.Module]]: | ||
| return self._target | ||
|
|
||
| @property | ||
|
|
@@ -160,7 +162,10 @@ def batch_index(self) -> Tuple: | |
|
|
||
| class CompositeLoss(BaseLoss): | ||
| def __init__( | ||
| self, loss_fn: Callable, name: str = "", target: nn.Module = [] | ||
| self, | ||
| loss_fn: Callable, | ||
| name: str = "", | ||
| target: Union[nn.Module, List[nn.Module]] = [], | ||
| ) -> None: | ||
| super(CompositeLoss, self).__init__(target) | ||
| self.__name__ = name | ||
|
|
@@ -499,6 +504,94 @@ def __call__(self, targets_to_values: ModuleOutputMapping) -> torch.Tensor: | |
| return _dot_cossim(self.direction, activations, cossim_pow=self.cossim_pow) | ||
|
|
||
|
|
||
| @loss_wrapper | ||
| class AngledNeuronDirection(BaseLoss): | ||
| """ | ||
| Visualize a direction vector with an optional whitened activation vector to | ||
| unstretch the activation space. Compared to the traditional Direction objectives, | ||
| this objective places more emphasis on angle by optionally multiplying the dot | ||
| product by the cosine similarity. | ||
|
|
||
| When cossim_pow is equal to 0, this objective works as a euclidean | ||
| neuron objective. When cossim_pow is greater than 0, this objective works as a | ||
| cosine similarity objective. An additional whitened neuron direction vector | ||
| can optionally be supplied to improve visualization quality for some models. | ||
|
|
||
| More information on the algorithm this objective uses can be found here: | ||
| https://github.com/tensorflow/lucid/issues/116 | ||
|
|
||
| This Lucid equivalents of this loss function can be found here: | ||
| https://github.com/tensorflow/lucid/blob/master/notebooks/ | ||
| activation-atlas/activation-atlas-simple.ipynb | ||
| https://github.com/tensorflow/lucid/blob/master/notebooks/ | ||
| activation-atlas/class-activation-atlas.ipynb | ||
|
|
||
| Like the Lucid equivalents, our implementation differs slightly from the | ||
| associated research paper. | ||
|
|
||
| Carter, et al., "Activation Atlas", Distill, 2019. | ||
| https://distill.pub/2019/activation-atlas/ | ||
| """ | ||
|
|
||
| def __init__( | ||
| self, | ||
| target: torch.nn.Module, | ||
| vec: torch.Tensor, | ||
| vec_whitened: Optional[torch.Tensor] = None, | ||
| cossim_pow: float = 4.0, | ||
| x: Optional[int] = None, | ||
| y: Optional[int] = None, | ||
| eps: float = 1.0e-4, | ||
| batch_index: Optional[int] = None, | ||
| ) -> None: | ||
| """ | ||
| Args: | ||
| target (nn.Module): A target layer instance. | ||
| vec (torch.Tensor): A neuron direction vector to use. | ||
| vec_whitened (torch.Tensor, optional): A whitened neuron direction vector. | ||
| cossim_pow (float, optional): The desired cosine similarity power to use. | ||
| x (int, optional): Optionally provide a specific x position for the target | ||
| neuron. | ||
| y (int, optional): Optionally provide a specific y position for the target | ||
| neuron. | ||
| eps (float, optional): If cossim_pow is greater than zero, the desired | ||
| epsilon value to use for cosine similarity calculations. | ||
| """ | ||
| BaseLoss.__init__(self, target, batch_index) | ||
| self.vec = vec.unsqueeze(0) if vec.dim() == 1 else vec | ||
| self.vec_whitened = vec_whitened | ||
| self.cossim_pow = cossim_pow | ||
| self.eps = eps | ||
| self.x = x | ||
| self.y = y | ||
| if self.vec_whitened is not None: | ||
| assert self.vec_whitened.dim() == 2 | ||
| assert self.vec.dim() == 2 | ||
|
|
||
| def __call__(self, targets_to_values: ModuleOutputMapping) -> torch.Tensor: | ||
| activations = targets_to_values[self.target] | ||
| activations = activations[self.batch_index[0] : self.batch_index[1]] | ||
| assert activations.dim() == 4 or activations.dim() == 2 | ||
| assert activations.shape[1] == self.vec.shape[1] | ||
| if activations.dim() == 4: | ||
| _x, _y = get_neuron_pos( | ||
| activations.size(2), activations.size(3), self.x, self.y | ||
| ) | ||
| activations = activations[..., _x, _y] | ||
|
|
||
| vec = ( | ||
| torch.matmul(self.vec, self.vec_whitened)[0] | ||
| if self.vec_whitened is not None | ||
| else self.vec | ||
| ) | ||
| if self.cossim_pow == 0: | ||
| return activations * vec | ||
|
|
||
| dot = torch.mean(activations * vec) | ||
| cossims = dot / (self.eps + torch.sqrt(torch.sum(activations ** 2))) | ||
|
There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. The code I've written follows after the implementation in the notebooks associated with the activation atlas papers:
They setup the objective calculations like this: The PyTorch objective merges these two objectives and follows what they do. There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. Thank you, @ProGamerGov for the reference! I think that it would be good to cite those notebooks in the code because it is in general confusing if the formal definitions vary from the implementation. To be honest. I don't quite understand why they explicitly defined and implemented that way. There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. @NarineK I'll add the references, and a note about how the reference code differs slightly from the implementation described in the paper! There was a problem hiding this comment. Choose a reason for hiding this commentThe reason will be displayed to describe this comment to others. Learn more. I also have yet to figure out why their implementation is slightly different than the paper, but I figured that I should follow the working reference implementation rather than the paper's equations for now! |
||
| return dot * torch.clamp(cossims, min=0.1) ** self.cossim_pow | ||
|
|
||
|
|
||
| @loss_wrapper | ||
| class TensorDirection(BaseLoss): | ||
| """ | ||
|
|
@@ -590,6 +683,47 @@ def __call__(self, targets_to_values: ModuleOutputMapping) -> torch.Tensor: | |
| return activations | ||
|
|
||
|
|
||
| def sum_loss_list( | ||
| loss_list: List, | ||
| to_scalar_fn: Callable[[torch.Tensor], torch.Tensor] = torch.mean, | ||
| ) -> CompositeLoss: | ||
| """ | ||
| Summarize a large number of losses without recursion errors. By default using 300+ | ||
| loss functions for a single optimization task will result in exceeding Python's | ||
| default maximum recursion depth limit. This function can be used to avoid the | ||
| recursion depth limit for tasks such as summarizing a large list of loss functions | ||
| with the built-in sum() function. | ||
|
|
||
| This function works similar to Lucid's optvis.objectives.Objective.sum() function. | ||
|
|
||
| Args: | ||
|
|
||
| loss_list (list): A list of loss function objectives. | ||
| to_scalar_fn (Callable): A function for converting loss function outputs to | ||
| scalar values, in order to prevent size mismatches. | ||
| Default: torch.mean | ||
|
|
||
| Returns: | ||
| loss_fn (CompositeLoss): A composite loss function containing all the loss | ||
| functions from `loss_list`. | ||
| """ | ||
|
|
||
| def loss_fn(module: ModuleOutputMapping) -> torch.Tensor: | ||
| return sum([to_scalar_fn(loss(module)) for loss in loss_list]) | ||
|
|
||
| name = "Sum(" + ", ".join([loss.__name__ for loss in loss_list]) + ")" | ||
| # Collect targets from losses | ||
| target = [ | ||
| target | ||
| for targets in [ | ||
| [loss.target] if not hasattr(loss.target, "__iter__") else loss.target | ||
| for loss in loss_list | ||
| ] | ||
| for target in targets | ||
| ] | ||
| return CompositeLoss(loss_fn, name=name, target=target) | ||
|
|
||
|
|
||
| def default_loss_summarize(loss_value: torch.Tensor) -> torch.Tensor: | ||
| """ | ||
| Helper function to summarize tensor outputs from loss functions. | ||
|
|
@@ -617,7 +751,9 @@ def default_loss_summarize(loss_value: torch.Tensor) -> torch.Tensor: | |
| "Alignment", | ||
| "Direction", | ||
| "NeuronDirection", | ||
| "AngledNeuronDirection", | ||
| "TensorDirection", | ||
| "ActivationWeights", | ||
| "sum_loss_list", | ||
| "default_loss_summarize", | ||
| ] | ||
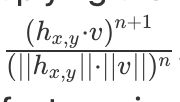
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
since cosine similarity is also seen as normalized dot product, it sounds a bit unclear what cosine similarity is multiplied to what dot product.
Uh oh!
There was an error while loading. Please reload this page.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
So, this is what
AngledNeuronDirectiondoes:And this is what
_dot_cossimdoes:There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thank you, @ProGamerGov! I saw that. I think the description of multiplying dot product by itself and normalizing it is a bit confusing. Is there any theoretical explanations for this.
Uh oh!
There was an error while loading. Please reload this page.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
@NarineK From the Activation Atlas paper, I think this might answer your question:
The Lucid Github issue lists a bunch of different feature visualization objective algorithms, including the one we are using (I copied the relevant part from the issue below):
Dot x Cosine Similarity
dot(x,y) * ceil(0.1, cossim(x,y))^nto avoid multiplying dot product by 0 or negative cosine similarity. Otherwise, you could end up in a situation where you maximize the opposite direction (because both dot and cossim are negative, and multiply to be positive) or get stuck because both are zero.There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Okay, I've added a link to the Lucid Github issue detailing the reasoning behind the loss algorithm!