-
Notifications
You must be signed in to change notification settings - Fork 2.1k
move width computation to setup_params()
#4416
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
Is this computation done per facet? |
|
No, globally. I'm not married to either but it seems we usually do these kind of computations globally |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
This seems safe to me, but I don't have strong sense of what the consequences are. But I can't imagine that you want different bar widths in different groups or facets.
|
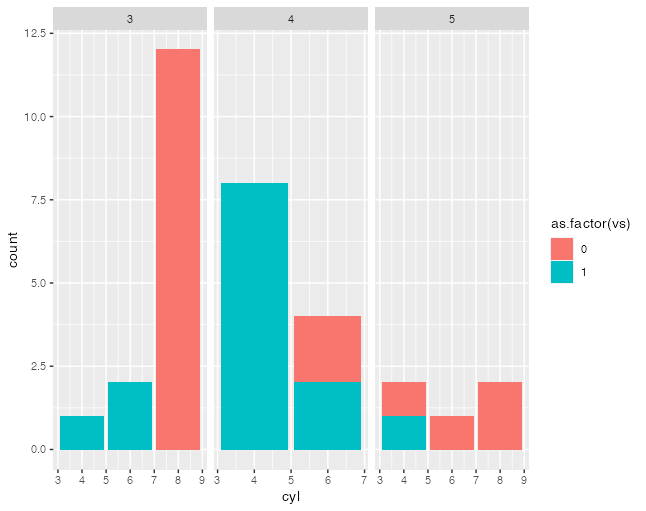
I think the case to check is a faceted plot where there are different numbers of bars and axes are set to free. I can't think of a dataset off the top of my head to make such a plot, but I have definitely encountered them. The current behavior is to adjust the bar width to fill up the available space. I'm not sure if that's good or bad, but we should at least check it. Such plots certainly do exist. If I have some time later today I'll try to make an example. |
library(ggplot2)
ggplot(mtcars,aes(cyl,fill=as.factor(vs)))+
geom_bar()+
facet_grid(~gear,scales="free" )
#> Warning: position_stack requires non-overlapping x intervals
#> Warning: position_stack requires non-overlapping x intervalsCreated on 2021-04-13 by the reprex package (v2.0.0) |
|
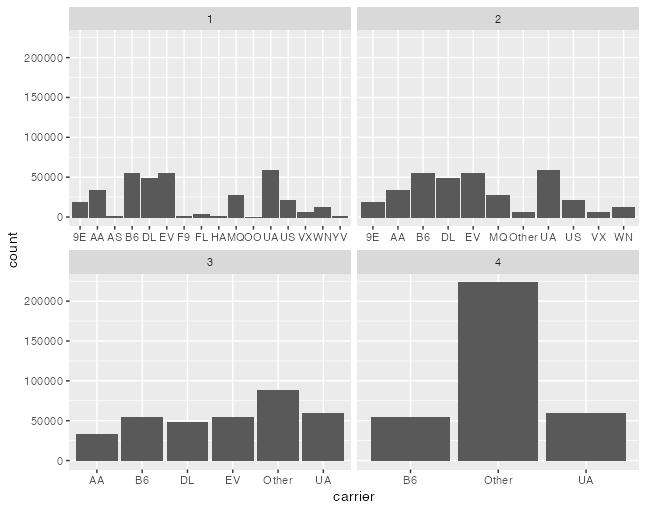
I meant something like this. Will it change? Also, notice how I have no strong opinion on whether bar widths should be the same or not in this type of a plot, I just wanted to point out that I've seen plots like this one, more than once. library(tidyverse)
library(nycflights13)
d1 <- flights %>%
select(carrier) %>%
mutate(group = 1)
d2 <- flights %>%
select(carrier) %>%
mutate(
carrier = as.character(fct_lump_n(carrier, n = 10)),
group = 2
)
d3 <- flights %>%
select(carrier) %>%
mutate(
carrier = as.character(fct_lump_n(carrier, n = 5)),
group = 3
)
d4 <- flights %>%
select(carrier) %>%
mutate(
carrier = as.character(fct_lump_n(carrier, n = 2)),
group = 4
)
d <- rbind(d1, d2, d3, d4)
d %>%
ggplot(aes(carrier)) +
geom_bar() +
facet_wrap(~group, scales = "free_x")d %>%
count(carrier, group) %>%
ggplot(aes(carrier, n)) +
geom_col() +
facet_wrap(~group, scales = "free_x")Created on 2021-04-13 by the reprex package (v1.0.0) |
|
@clauswilke the output doesn't change because the width is encoded in the scale domain, i.e. a width of 0.9 seems bigger in a panel with fewer categories |
|
For completeness, this is @smouksassi's example rendered with this patch |



Fix #2047
This PR makes sure that width is calculated globally to avoid surprise behaviour outlined in #2047. There is a chance that this will break someones code, but I think it is pretty low