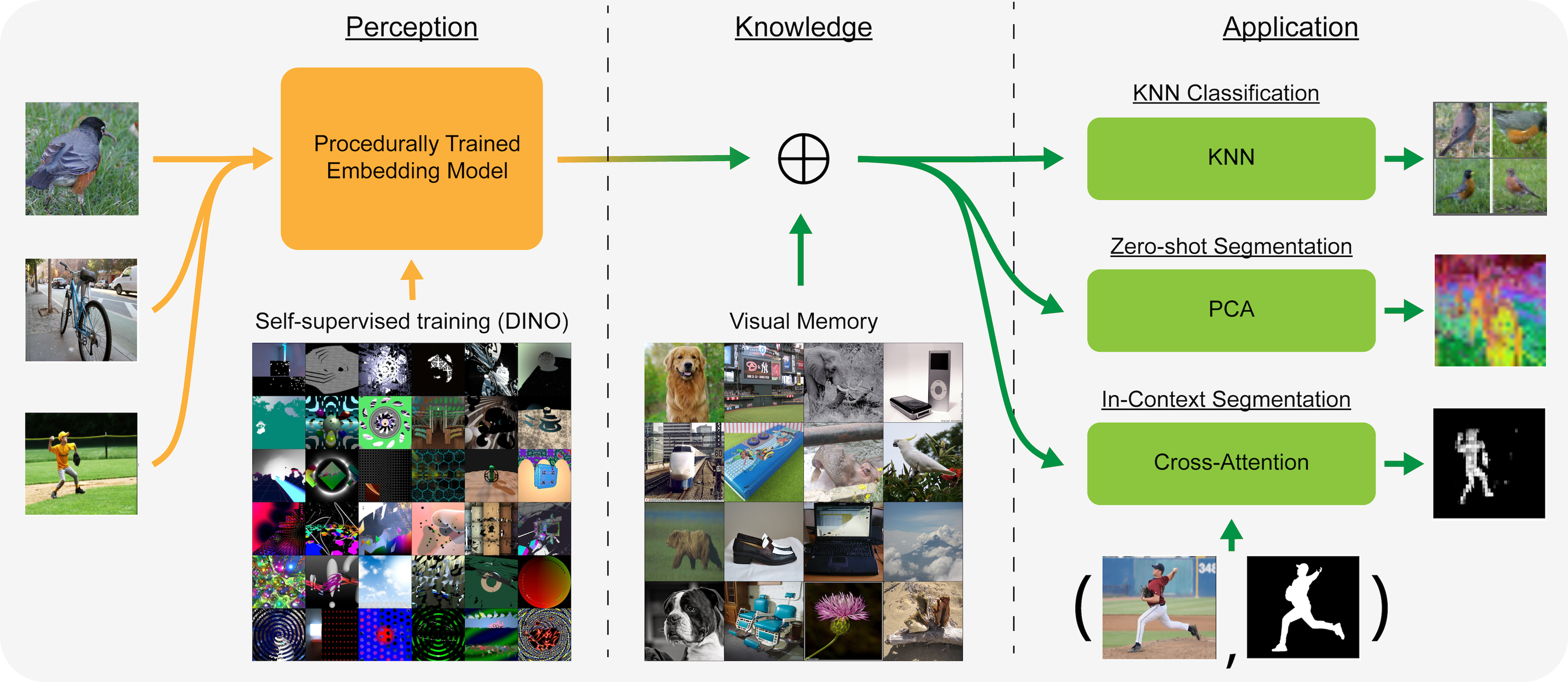
Welcome to the repository for "Separating Knowledge with Procedural Data"! Here are the instructions for reproducing all trainings and evaluations in the paper.
[Project page] [Paper]
conda create -n separating_knowledge python=3.9.20
conda activate separating_knowledge
pip install torch==2.4.1 torchaudio==2.4.1 torchvision==0.19.1
pip install importlib-metadata
pip install opencv-python
pip install timm
pip install matplotlib
pip install pandas
pip install pykeops
pip install seaborn
pip install medmnist
pip install pycocotools
pip install scikit-learn
Follow each subsection instructions, starting each subsection in the main directory.
See their respective websites for instructions ImageNet and Places, then symlink to ./data.
cd download_data_scripts
./download_stylegan.sh YOUR_DATASETS_FOLDER_HERE
./download_shaders.sh YOUR_DATASETS_FOLDER_HERE
mkdir data
cd data
mkdir imagenet
mkdir places
mkdir shaders_mixup
mkdir shaders
mkdir stylegan
ln -s PATH_TO_IMAGENET/train imagenet/train
ln -s PATH_TO_IMAGENET/val imagenet/val
ln -s PATH_TO_PLACES/train places/train
ln -s PATH_TO_SHADERS_MIXUP/train shaders_mixup/train
ln -s PATH_TO_SHADERS/train shaders/train
ln -s PATH_TO_STYLEGAN/train stylegan/train
cd data_generation
# Shaders KML
./shaders_kml.sh PATH_TO_DATASET_FOLDER
mkdir ../data/shaders_kml
ln -s PATH_TO_SHADERS_KML/train ../data/shaders_kml/train
# Shaders KML Mixup
./shaders_kml_mixup.sh PATH_TO_DATASET_FOLDER
mkdir ../data/shaders_kml_mixup
ln -s PATH_TO_SHADERS_KML_MIXUP/train ../data/shaders_kml_mixup/train
See their respective websites for instructions CUB, Flowers102, and Food, then symlink to ./data.
See the website MedicalMNIST for instructions, download in 224x244 resolution and then symlink to ./data.
See their respective websites for instructions COCO, Ade20k, and Pascal, then symlink to ./data.
cd dino
./scripts/train_imagenet.sh
./scripts/train_places.sh
./scripts/train_shaders_kml_mixup.sh
./scripts/train_shaders_kml.sh
./scripts/train_shaders_mixup.sh
./scripts/train_shaders.sh
./scripts/train_stylegan.sh
cd dino
# ENCODER_NAME=imagenet, shaders_kml_mixup, etc.
./scripts/evals/eval_knn_cub.sh ENCODER_NAME # ENCODER_NAME=imagenet, shaders_kml_mixup, etc.
./scripts/evals/eval_knn_flowers.sh ENCODER_NAME # ENCODER_NAME=imagenet, shaders_kml_mixup, etc.
./scripts/evals/eval_knn_food.sh ENCODER_NAME # ENCODER_NAME=imagenet, shaders_kml_mixup, etc.
cd dino
# ENCODER_NAME=imagenet, shaders_kml_mixup, etc.
# DATASET_NAME=bloodmnist, breastmnist, dermamnist, octmnist, organamnist, organcmnist, organsmnist, pathmnist, pneumoniamnist, tissuemnist
./scripts/evals/eval_knn_medicalmnist.sh ENCODER_NAME DATASET_NAME
cd dino
# ENCODER_NAME=imagenet, shaders_kml_mixup, etc.
python dump_coco_features.py --pretrained_weights ./encoders/ENCODER_NAME/checkpoint.pth
Open notebooks notebook_figures/figures_segmentation_zeroshot.ipynb, notebook_figures/figures_segmentation_incontext.ipynb, and notebook_figures/figures_segmentation_knn.ipynb.